

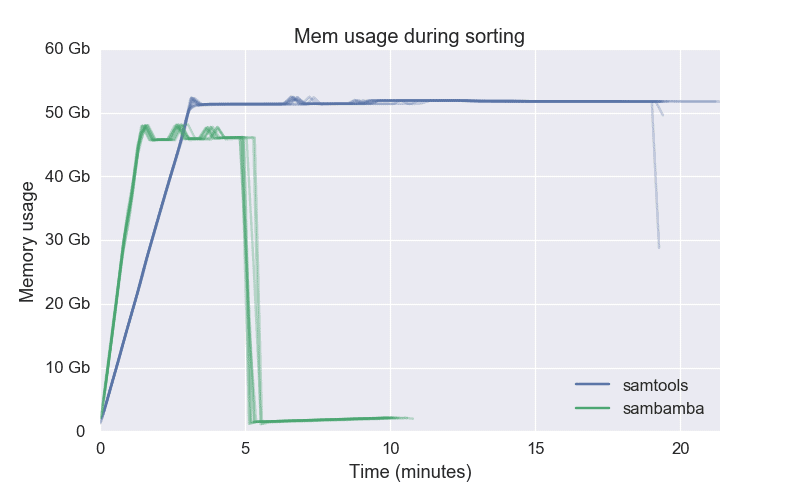
OpenMP task parallelism to enhance concurrency and memory optimization techniques were employed in both SAMtools and the underlying library HTSlib. This paper presents a case study on the performance characterization and optimization of BAM sorting with SAMtools. As a result, BAM-file sorting can be a bottleneck in genomics workflows. This can be computationally- and I/O-intensive: BAM files can be many gigabytes in size, and may need to be decompressed before sorting and compressed afterwards. A common use of SAMtools is to sort the standard Binary Alignment/Map (BAM) format emitted by many sequence aligners. The statistics options signficantly reduce the speed at whichĭeduplication is performed and increase the memory usage.SAMtools is a suite of tools that is widely-used in genomics workflows for post-processing sequence alignment data from large high-throughput sequencing data sets. UMIs is 1, whereas in the random null (in the final column) we would Looking at the third row, we see that there areġ167 positions where the average edit distance between Where every UMI is assumed to represent an independent molecule in theīiological sample. The last twoĬolumns the same, but for the naive unique deduplication method In the overall usage of particular UMI sequences). The secondĬolumn is what we would expect to see if UMIs were randomlyĭistributed between mapping locations (taking into account any biases Line we see that there are 2 bases in the genome where the averageĮdit distance between the UMIs found at that base is 1. With the directional-adjacency method (the default). The first two columns show the distribution of average edit distancesīetween UMIs found at a single base in the genome after deduplication The content of this file after running the above will look something like: directional-adjacency

One of the most interesting is theĭistribution of edit distances (here named deduplicated_edit_distance.tsv). The -output-stats option is optional, but selecting it will provide a range $ umi_tools dedup -I example.bam -output-stats=deduplicated -S deduplicated.bam Will need indexing with samtools index before use. Have the correct indexs install etc, we have provided a BAM file of The ability of folks to follow along without having to worry if they
It is also the least interesting for us here. The most computationally intensive part of this is the middle part. Have adaptor trimmed and filtered the data to reduce its size.Įxtract UMI from raw reads -> map reads -> deduplicate reads based on UMIs Theĭata used comes from one of the control replicates from Mueller-Mcnicoll et al, Use the UMI-tools package to process data with UMIs added to them. The following steps will guide you through a short example of how to If you want to apply UMI-tools in a single cell RNA-Seq analysis, please see the Single cell tutorial
This quick start guide uses an iCLIP dataset as an example.


 0 kommentar(er)
0 kommentar(er)
